Identification of hub biomarkers and immune-related pathways associated with sepsis
-
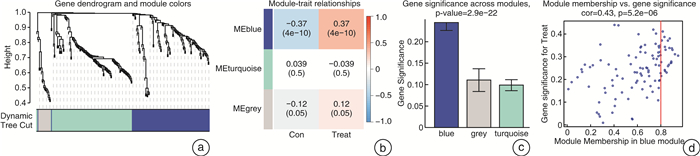
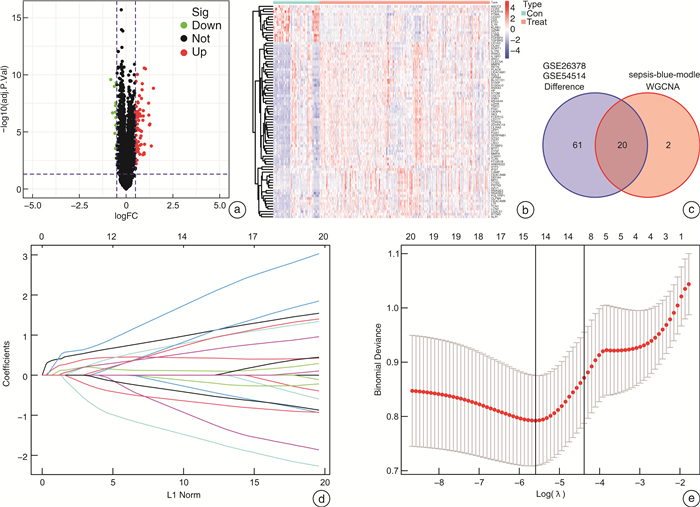
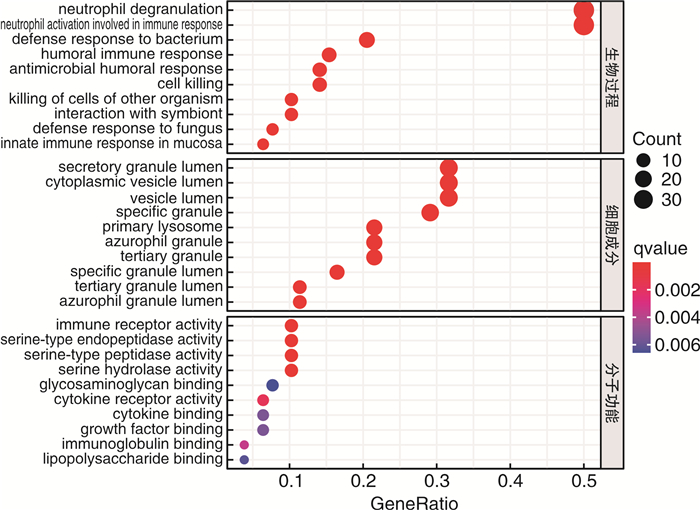
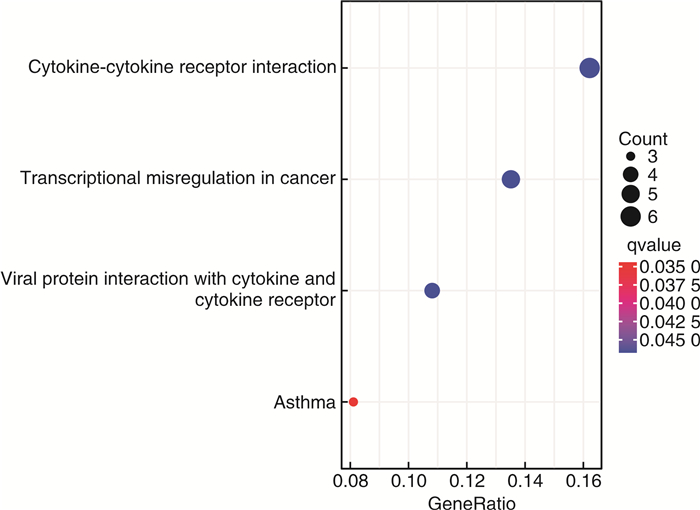
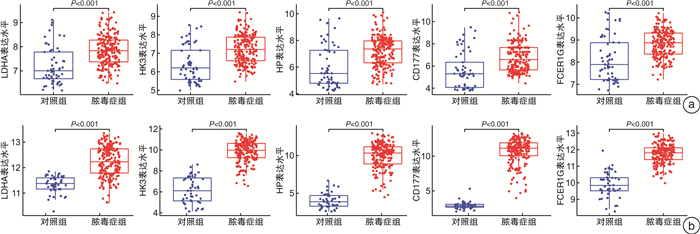
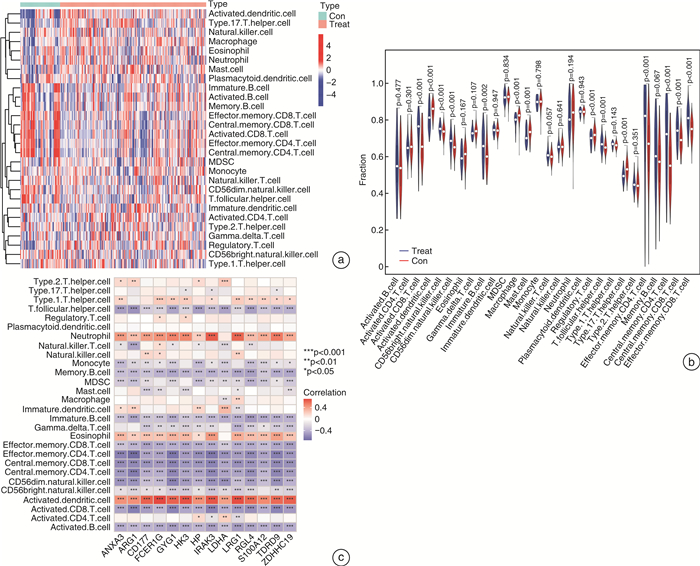
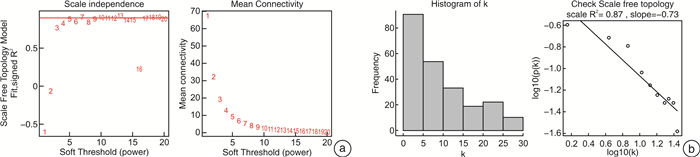
摘要: 目的 确定参与脓毒症的关键生物标志物和免疫相关途径及其与免疫细胞浸润的关系。方法 在GEO网站下载GSE26378、GSE54514和GSE66099数据集。通过差异基因表达分析、加权基因共表达网络分析(WGCNA)、最小绝对值选择与收缩算子(LASSO)回归分析等方法挖掘脓毒症核心标志物,通过基因本体分析(GO)、京都基因与基因组百科全书分析(KEGG)、基因集富集分析(GSEA)对差异基因(DEGs)进行表型分析。然后,采用受试者工作特征曲线(ROC)验证核心标志物对脓毒症诊断的准确性。最后,采用单样本基因集富集分析(ssGSEA),分析28个免疫细胞在表达谱中的浸润水平及其与核心基因标记的关系。结果 共筛选出81个差异基因。通过WGCNA分析获得3个共表达模块,其中蓝色模块与脓毒症的相关性最高。结合差异基因表达分析,共获得20个交叉基因。随后,通过LASSO回归分析,5个核心基因(LDHA、HK3、HP、CD177和FCER1G)被鉴定为脓毒症的潜在生物标志物。免疫浸润结果显示骨髓源性抑制细胞(MDSC)、单核细胞、活化树突状细胞、中性粒细胞和巨噬细胞之间的关系最为显著。ROC曲线分析显示了5个核心基因的主要诊断价值。差异基因功能富集分析显示,核心基因主要在免疫和炎症通路中增强。结论 B细胞和单核细胞与脓毒症的发病密切相关。核心基因LDHA、HK3、HP、CD177和FCER1G可能通过免疫相关信号通路参与脓毒症的进展。
-
关键词:
- 脓毒症 /
- 免疫细胞浸润 /
- 免疫相关通路 /
- 最小绝对值选择与收缩算子 /
- 加权基因共表达网络分析
Abstract: Objective To identify key biomarkers and immune-related pathways involved in the progression of sepsis and their relationship with immune cell infiltration.Methods The GSE26378, GSE54514, and GSE66099 datasets were downloaded from the Gene Expression Omnibus database. Hub markers for sepsis were mined through differential expression analysis, weighted gene co-expression network analysis(WGCNA), and least absolute shrinkage and selection operator(LASSO) analysis, and differential genes were characterized by Gene Ontology analysis, Kyoto Encyclopedia of Genes and Genomes(KEGG) analysis, and Gene Set Enrichment Analysis(GSEA). Then, the receiver operating characteristic curve(ROC) was used in verifying the accuracy of the hub markers for sepsis diagnosis. Finally, single-sample gene set enrichment analysis was used in analyzing the infiltration levels of 28 immune cells in the expression profile and their relationship to hub gene markers.Results A total of 81 differential genes were screened. Three co-expression modules were obtained through WGCNA; in which, the blue module had the highest correlation with sepsis. A total of 20 intersecting genes were acquired by combining differential genes. Subsequently, five hub genes were identified by LASSO analysis as potential biomarkers for sepsis. Immune infiltration results revealed the most significant relationship among myeloid-derived suppressor cells(MDSC), monocytes, activated dendritic cells, neutrophils, and macrophages. ROC curve analysis demonstrated the prime diagnostic value of the five hub genes. Functional enrichment analysis of differential genes showed that hub genes were mainly enhanced in immune and inflammatory pathways.Conclusion B cells and monocytes were closely associated with the pathogenesis of sepsis. The hub genes lactate dehydrogenase A, hexokinase 3, haptoglobin, CD177, and FCER1G may be involved in the progression of sepsis through immune-related signal pathways. -
-
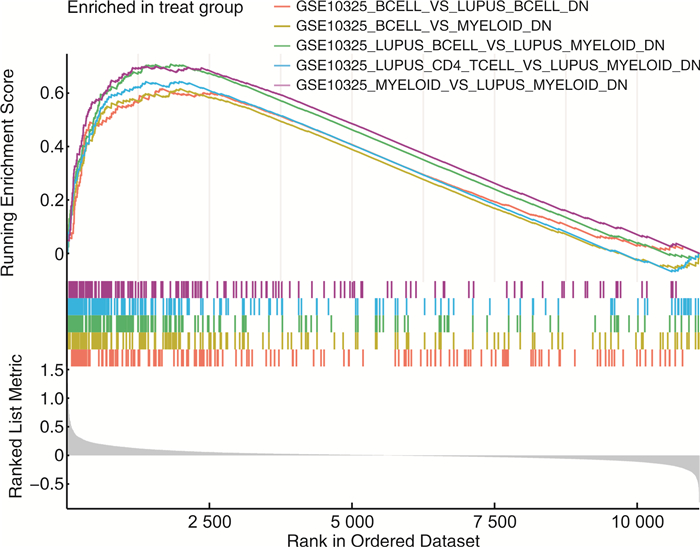
表 1 GSEA中DEGs富集的前5个重要免疫信号
基因集名称 NES P NOM p-val FDR q-val GSE10325_BCELL_VS_LUPUS_BCELL_DN 2.189 < 0.001 < 0.001 < 0.001 GSE10325_BCELL_VS_MYELOID_DN 2.191 < 0.001 < 0.001 < 0.001 GSE10325_LUPUS_BCELL_VS_LUPUS_MYELOID_DN 2.524 < 0.001 < 0.001 < 0.001 GSE10325_LUPUS_CD4_TCELL_VS_LUPUS_MYELOID_DN 2.286 < 0.001 < 0.001 < 0.001 GSE10325_MYELOID_VS_LUPUS_MYELOID_DN 2.486 < 0.001 < 0.001 < 0.001 -
[1] Torres LK, Pickkers P, van der Poll T. Sepsis-induced immunosuppression[J]. Annu Rev Physiol, 2022, 84: 157-181. doi: 10.1146/annurev-physiol-061121-040214
[2] 陈正钢, 刘励军. 急诊脓毒症患者早期筛查生物标志物的研究现状与展望[J]. 临床急诊杂志, 2023, 24(2): 99-104. doi: 10.13201/j.issn.1009-5918.2023.02.010 https://lcjz.whuhzzs.com/article/doi/10.13201/j.issn.1009-5918.2023.02.010
[3] Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis[J]. BMC Bioinformatics, 2008, 9: 559. doi: 10.1186/1471-2105-9-559
[4] Duan JB, Soussen C, Brie D, et al. Generalized LASSO with under-determined regularization matrices[J]. Signal Processing, 2016, 127: 239-246. doi: 10.1016/j.sigpro.2016.03.001
[5] Sun CS, Han JT, Bai YX, et al. Neuropeptides as the shared genetic crosstalks linking periodontitis and major depression disorder[J]. Dis Markers, 2021, 2021: 3683189. http://www.xueshufan.com/publication/3207091808
[6] Boomer JS, Green JM, Hotchkiss RS. The changing immune system in sepsis: is individualized immuno-modulatory therapy the answer?[J]. Virulence, 2014, 5(1): 45-56. doi: 10.4161/viru.26516
[7] Boyd JH, Russell JA, Fjell CD. The meta-genome of sepsis: host genetics, pathogens and the acute immune response[J]. J Innate Immun, 2014, 6(3): 272-283. doi: 10.1159/000358835
[8] Kolaczkowska E, Kubes P. Neutrophil recruitment and function in health and inflammation[J]. Nat Rev Immunol, 2013, 13(3): 159-175. doi: 10.1038/nri3399
[9] Santopolo G, Clemente A, Aranda M, et al. Colorimetric detection of sepsis-derived hyperdegranulation with plasmonic nanosensors[J]. ACS Sens, 2021, 6(12): 4443-4450. doi: 10.1021/acssensors.1c01884
[10] Edwards YH, Povey S, Levan KM, et al. Locus determining the human sperm-specific lactate dehydrogenase, LDHC, is syntenic with LDHA[J]. Dev Genet, 1987, 8(4): 219-232. doi: 10.1002/dvg.1020080406
[11] Pan TT, Sun SQ, Chen Y, et al. Immune effects of PI3K/akt/HIF-1α-regulated glycolysis in polymorphonuclear neutrophils during sepsis[J]. Crit Care, 2022, 26(1): 29. doi: 10.1186/s13054-022-03893-6
[12] Smith JA, Stallons LJ, Schnellmann RG. Renal cortical hexokinase and pentose phosphate pathway activation through the EGFR/Akt signaling pathway in endotoxin-induced acute kidney injury[J]. Am J Physiol Renal Physiol, 2014, 307(4): F435-F444. doi: 10.1152/ajprenal.00271.2014
[13] Andersen CBF, Stødkilde K, Søderup KL, et al. Haptoglobin[J]. Antioxid Redox Signal, 2017, 26(14): 814-831. doi: 10.1089/ars.2016.6793
[14] Raju SM, Kumar AP, Yadav AN, et al. Haptoglobin improves acute phase response and endotoxin tolerance in response to bacterial LPS[J]. Immunol Lett, 2019, 207: 17-27. doi: 10.1016/j.imlet.2019.01.002
[15] Wolff JC, Goehring K, Heckmann M, et al. Sex-dependent up regulation of CD 177-specific mRNA expression in cord blood due to different stimuli[J]. Transfusion, 2006, 46(1): 132-136. doi: 10.1111/j.1537-2995.2005.00676.x
[16] Demaret J, Venet F, Plassais J, et al. Identification of CD177 as the most dysregulated parameter in a microarray study of purified neutrophils from septic shock patients[J]. Immunol Lett, 2016, 178: 122-130. doi: 10.1016/j.imlet.2016.08.011
[17] Liang Y, Wang P, Zhao M, et al. Demethylation of the FCER1G promoter leads to FcεRI overexpression on monocytes of patients with atopic dermatitis[J]. Allergy, 2012, 67(3): 424-430. doi: 10.1111/j.1398-9995.2011.02760.x
[18] Zou M, Su XY, Wang LY, et al. The molecular mechanism of multiple organ dysfunction and targeted intervention of COVID-19 based on time-order transcriptomic analysis[J]. Front Immunol, 2021, 12: 729776. doi: 10.3389/fimmu.2021.729776
[19] de Zuani M, Hortová-Kohoutková M, Andrejčinová I, et al. Human myeloid-derived suppressor cell expansion during sepsis is revealed by unsupervised clustering of flow cytometric data[J]. Eur J Immunol, 2021, 51(7): 1785-1791. doi: 10.1002/eji.202049141
[20] Zhang WY, Fang XZ, Gao CG, et al. MDSCs in sepsis-induced immunosuppression and its potential therapeutic targets[J]. Cytokine Growth Factor Rev, 2023, 69: 90-103. doi: 10.1016/j.cytogfr.2022.07.007
[21] Ren ZF, Tang HR, Wan LJ, et al. Swertianolin ameliorates immune dysfunction in sepsis via blocking the immunosuppressive function of myeloid-derived suppressor cells[J]. Eur J Histochem, 2021, 65(3): 3292. http://www.xueshufan.com/publication/3198826028
[22] Hausfater P, Robert Boter N, Morales Indiano C, et al. Monocyte distribution width(MDW)performance as an early sepsis indicator in the emergency department: comparison with CRP and procalcitonin in a multicenter international European prospective study[J]. Crit Care, 2021, 25(1): 227. doi: 10.1186/s13054-021-03622-5
[23] Chen LS, Zhao YF, Lai DM, et al. Neutrophil extracellular traps promote macrophage pyroptosis in sepsis[J]. Cell Death Dis, 2018, 9: 597. doi: 10.1038/s41419-018-0538-5
[24] Liu WZ, Liu TY, Zheng YJ, et al. Metabolic reprogramming and its regulatory mechanism in sepsis-mediated inflammation[J]. J Inflamm Res, 2023, 16: 1195-1207. doi: 10.2147/JIR.S403778
[25] Liu D, Huang SY, Sun JH, et al. Sepsis-induced immunosuppression: mechanisms, diagnosis and current treatment options[J]. Mil Med Res, 2022, 9(1): 56.
[26] Pei F, Yao RQ, Ren C, et al. Expert consensus on the monitoring and treatment of sepsis-induced immunosuppression[J]. Mil Med Res, 2022, 9(1): 74.
-



 下载:
下载: