AI pathogen type discrimination model for pneumonia based on multi-dimensional clinical data etiology
-
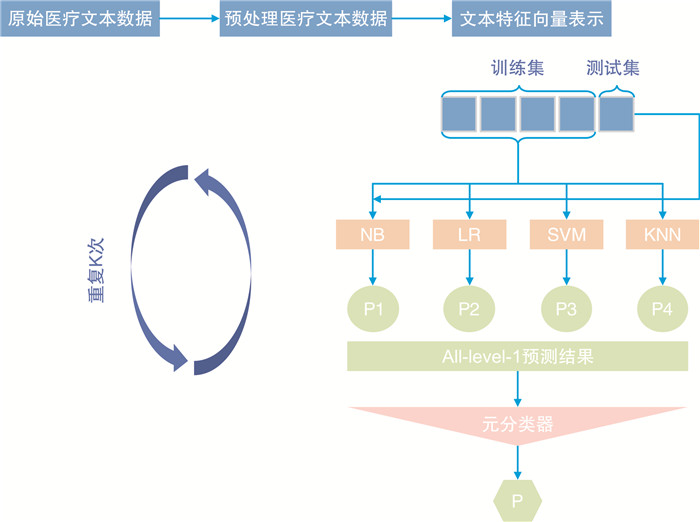
摘要: 目的 基于肺炎患者的临床资料,建立肺炎人工智能(artificial intelligence,AI)病原学类型判别模型,预测肺炎的责任病原体,帮助临床医生选择合适的抗感染治疗方案。方法 回顾性地收集了北京天坛医院急诊科与呼吸科在2018年1月—2020年12月收治的197例肺炎患者的临床资料。选取158例(80%)患者资料作为建模组,构建肺炎AI病原学类型判别模型,39例(20%)作为验证组,验证模型的预测效果。同时,将验证组预测结果与20名急诊科医师的病原学诊断结果进行对比。结果 基于多维度临床数据构建的肺炎AI病原学类型判别模型的病原学验证精度为94.87%。20名急诊科医师病原学诊断的准确率分别为7.69%、15.38%、10.26%、10.26%、15.38%、17.95%、12.82%、10.26%、25.64%、17.95%、7.69%、5.13%、12.80%、20.51%、17.95%、7.69%、28.21%、12.82%、23.08%、15.38%,该模型的验证精度高于临床医师病原学诊断的平均准确率(94.87% vs 14.74%)。结论 借助既往肺炎患者的临床资料,本研究创建了基于多维度临床数据的肺炎AI病原学类型判别模型,该模型可用于早期预测肺炎患者的责任病原体,为临床医生早期制定经验性抗感染治疗方案提供参考。受限于样本量,本模型的临床价值有待进一步研究。Abstract: Objective To establish an artificial intelligence (AI) model for pathogen discrimination in pneumonia patients using their medical records. The model aims to predict the causative pathogens of pneumonia and assist clinical physicians in selecting appropriate antimicrobial treatment strategies.Methods Retrospective medical records of 197 pneumonia patients admitted to the Emergency and Respiratory Departments of Beijing Tiantan Hospital from January 2018 to December 2020 were collected. Among these, data from 158 patients (80%) were selected to build the pneumonia AI pathogen discrimination model, while data from 39 patients (20%) were used for model validation. The predictive results of the validation group were also compared with the pathogen diagnoses made by twenty emergency department physicians.Results The AI pathogen discrimination model, based on multi-dimensional clinical data, achieved a pathogen validation accuracy of 94.87%. In contrast, the accuracy of pathogen diagnosis by the twenty emergency department physicians ranged from 5.13% to 28.21%, with an average accuracy of 14.74%. Therefore, the model's validation accuracy outperformed the average accuracy of clinical physician pathogen diagnoses (94.87% vs 14.74%).Conclusion By utilizing historical medical records of pneumonia patients, this study successfully developed an AI pathogen discrimination model based on multi-dimensional clinical data. The model shows promise in early prediction of pneumonia pathogens and provides valuable references for clinical physicians in selecting empirical antimicrobial treatment strategies. However, due to limitations in sample size, further research is warranted to explore the full clinical potential of this model.
-
Key words:
- pneumonia /
- multi-dimensional clinical data /
- aetiology /
- artificial intelligence
-
-
表 1 建模组与验证组肺炎患者的临床特征比较
临床资料 建模组
(158例)验证组
(39例)P 年龄/岁 52(42.39~56.42) 66(54.89~68.34) 0.060 性别/例(%) 0.820 男 90(56.96) 23(58.87) 女 68(43.04) 16(41.03) 病史/例(%) 受凉/淋雨 14(8.86) 6(15.38) 0.227 咳嗽 120(75.95) 37(94.87) 0.109 咳痰 103(65.19) 36(96.31) 0.101 乏力 4(2.53) 10(25.64) 0.071 胸闷/憋气 44(27.85) 25(64.10) 0.063 呼吸困难 12(7.59) 4(10.26) 0.393 发热 65(41.14) 22(56.41) 0.085 寒战 11(6.96) 3(7.69) 0.550 头痛 4(2.53) 5(12.82) 0.016 咽痛 6(3.80) 2(5.13) 0.494 肌痛 8(5.06) 0 0.165 化验检查 白细胞/(×109/L) 12.36(4.38) 12.09(5.00) 0.015 中性粒细胞/(×109/L) 8.1(3.74) 9.66(4.89) 0.216 PCT/(ng/mL) 0.32(0.62~2.13) 0.31(0.51~3.63) 0.489 CRP/(mg/L) 90.44(79.14) 110.18(89.24) 0.392 痰细菌学检测结果/例(%) 支原体 18(11.39) 4(10.26) 0.551 军团菌 8(5.06) 3(7.69) 0.376 肺炎链球菌 11(6.96) 2(5.13) 0.506 大肠埃希菌 8(5.06) 2(5.13) 0.624 铜绿假单胞菌 16(10.13) 7(17.94) 0.140 肺炎克雷伯菌 23(14.56) 7(17.94) 0.598 流感嗜血杆菌 7(4.43) 1(2.56) 0.506 鲍曼不动杆菌 32(20.25) 6(15.38) 0.490 金黄色葡萄球菌 23(14.56) 5(12.82) 0.781 嗜麦芽窄食单胞菌 12(7.59) 2(5.13) 0.450 表 2 建模组与验证组肺炎患者的影像学特征比较
例(%) 影像学特征 建模组
(158例)验证组
(39例)P 双肺感染 68(43.04) 21(53.85) 0.225 磨玻璃影 16(23.53) 3(14.29) 0.282 斑片影 8(11.76) 5(23.81) 0.172 炎性改变 44(64.71) 13(61.90) 0.815 合并支扩 36(52.94) 3(14.29) 0.002 合并肺气肿 24(79.41) 11(52.38) 0.161 合并肺大疱 19(27.94) 4(19.05) 0.306 合并气胸 5(7.35) 1(0) 0.565 合并胸腔积液 32(47.06) 8(38.10) 0.470 合并支气管壁增厚 18(26.47) 6(28.57) 0.850 单肺感染 90(56.96) 18(46.15) 0.225 单肺多叶 54(60.00) 11(61.11) 0.930 磨玻璃影 10(18.52) 1(9.09) 0.401 斑片影 6(11.11) 1(9.09) 0.662 炎性改变 38(70.37) 9(81.82) 0.357 合并支扩 12(22.22) 2(18.18) 0.562 合并肺气肿 17(31.48) 1(9.09) 0.124 合并肺大疱 7(12.96) 0 0.254 合并气胸 3(5.56) 0 0.568 合并胸腔积液 19(35.19) 3(27.27) 0.449 合并支气管壁增厚 24(44.44) 4(36.36) 0.441 单肺单叶 36(40.00) 7(38.89) 0.930 磨玻璃影 8(22.22) 2(28.57) 0.524 斑片影 2(5.56) 0 0.698 炎性改变 26(72.22) 5(71.43) 0.644 合并支扩 3(8.33) 0 0.579 合并肺气肿 8(22.22) 0 0.209 合并肺大疱 2(5.56) 0 0.698 合并气胸 1(0) 0 0.837 合并胸腔积液 2(5.56) 0 0.698 合并支气管壁增厚 15(41.67) 4(57.14) 0.365 表 3 建模组内部验证结果
肺炎病原体 金标准检测个数 模型输出结果/个 准确率/% 错误输出病原类型 正确 错误 支原体 18 18 0 100.00 - 军团菌 8 8 0 100.00 - 肺炎链球菌 11 11 0 100.00 - 金黄色葡萄球菌 23 22 1 95.65 鲍曼不动杆菌 大肠埃希菌 8 8 0 100.00 - 铜绿假单胞菌 16 15 1 93.75 肺炎克雷伯菌 肺炎克雷伯菌 23 22 1 95.65 鲍曼不动杆菌 流感嗜血杆菌 7 7 0 100.00 - 鲍曼不动杆菌 32 30 2 93.75 肺炎克雷伯菌×2 嗜麦芽窄食单胞菌 12 12 0 100.00 - 总体 158 153 5 96.83 - 注:肺炎克雷伯菌×2表示2例错误输出病原类型均为肺炎克雷伯菌。 表 4 验证组验证结果
肺炎病原体 金标准检测个数 模型输出结果/个 准确率/% 错误输出病原类型 正确 错误 支原体 4 4 0 100.00 - 军团菌 3 3 0 100.00 - 肺炎链球菌 2 2 0 100.00 - 金黄色葡萄球菌 5 5 0 100.00 - 大肠埃希菌 2 2 0 100.00 - 铜绿假单胞菌 7 7 0 100.00 - 肺炎克雷伯菌 7 7 0 100.00 - 流感嗜血杆菌 1 0 1 0 大肠埃希菌 鲍曼不动杆菌 6 5 1 83.33 肺炎克雷伯菌 嗜麦芽窄食单胞菌 2 2 0 100.00 - 总体 39 37 2 94.87 - 表 5 AI病原学判别模型判别结果与20名医师诊断结果
分组 诊断正确/个 诊断错误/个 正确率/% P AI模型组 37 2 94.87 医师1 3 36 7.69 <0.01 医师2 6 33 15.38 <0.01 医师3 4 35 10.26 <0.01 医师4 4 35 10.26 <0.01 医师5 6 33 15.38 <0.01 医师6 7 32 17.95 <0.01 医师7 5 34 12.82 <0.01 医师8 4 35 10.26 <0.01 医师9 10 29 25.64 <0.01 医师10 7 32 17.95 <0.01 医师11 3 36 7.69 <0.01 医师12 2 37 5.13 <0.01 医师13 5 34 12.82 <0.01 医师14 8 31 20.51 <0.01 医师15 7 32 17.95 <0.01 医师16 3 36 7.69 <0.01 医师17 11 28 28.21 <0.01 医师18 5 34 12.82 <0.01 医师19 9 30 23.08 <0.01 医师20 6 33 15.38 <0.01 -
[1] Scheld WM. Developments in the pathogenesis, diagnosis and treatment of nosocomial pneumonia[J]. Surg Gynecol Obstet, 1991, 172(Suppl): 42-53.
[2] Heron M. Deaths: leading causes for 2015[J]. Natl Vital Stat Rep, 2017, 66(5): 1-76.
[3] Chongthanadon B, Thirawattanasoot N, Ruangsomboon O. Clinical factors associated with in-hospital mortality in elderly versus non-elderly pneumonia patients in the emergency department[J]. BMC Pulm Med, 2023, 23(1): 330. doi: 10.1186/s12890-023-02632-z
[4] Hespanhol V, Bárbara C. Pneumonia mortality, comorbidities matter?[J]. Pulmonology, 2020, 26(3): 123-129. doi: 10.1016/j.pulmoe.2019.10.003
[5] Schöll N, Rohde GGU. Ambulant erworbene Pneumonie bei älteren Menschen [Community-acquired Pneumonia in the Elderly][J]. Pneumologie, 2019, 73(10): 605-616. doi: 10.1055/a-0835-1943
[6] 邹晓辉, 曹彬. 呼吸道感染病原学诊断年度进展2021[J]. 中华结核和呼吸杂志, 2022, 45(1): 78-82.
[7] 中华医学会检验医学分会. 高通量宏基因组测序技术检测病原微生物的临床应用规范化专家共识[J]. 中华检验医学杂志, 2020, 43(12): 1181-1195. doi: 10.3760/cma.j.cn114452-20200903-00704
[8] Modi AR, Kovacs CS. Hospital-acquired and ventilator-associated pneumonia: Diagnosis, management, and prevention[J]. Cleve Clin J Med, 2020, 87(10): 633-639. doi: 10.3949/ccjm.87a.19117
[9] Fleuren LM, Klausch TLT, Zwager CL, et al. Machine learning for the prediction of sepsis: a systematic review and meta-analysis of diagnostic test accuracy[J]. Intensive Care Med, 2020, 46(3): 383-400. doi: 10.1007/s00134-019-05872-y
[10] 潘丽艳, 梁会营. 基于深度学习的儿童肺炎病原学类型判别模型[J]. 中国数字医学, 2019, 14(3): 59-61, 110. https://www.cnki.com.cn/Article/CJFDTOTAL-YISZ201903021.htm
[11] Li YY, Zhang ZY, Dai C, et al. Accuracy of deep learning for automated detection of pneumonia using chest X-Ray images: a systematic review and meta-analysis[J]. Comput Biol Med, 2020, 123: 103898. doi: 10.1016/j.compbiomed.2020.103898
[12] Heckerling PS, Gerber BS, Tape TG, et al. Prediction of community-acquired pneumonia using artificial neural networks[J]. Med Decis Making, 2003, 23(2): 112-121. doi: 10.1177/0272989X03251247
[13] Kermany DS, Goldbaum M, Cai WJ, et al. Identifying medical diagnoses and treatable diseases by image-based deep learning[J]. Cell, 2018, 172(5): 1122-1131.e9. doi: 10.1016/j.cell.2018.02.010
[14] Hwang EJ, Park S, Jin KN, et al. Development and validation of a deep learning-based automated detection algorithm for major thoracic diseases on chest radiographs[J]. JAMA Netw Open, 2019, 2(3): e191095. doi: 10.1001/jamanetworkopen.2019.1095
[15] Stephen O, Sain M, Maduh UJ, et al. An efficient deep learning approach to pneumonia classification in healthcare[J]. J Healthc Eng, 2019, 2019: 4180949.
-



 下载:
下载: